Nano LC in proteomic analysis
Remco Swart, Thermo Scientific, NetherlandsAbstract Proteomics aims to characterize the entire complement of proteins expressed in a cell, tissue, or organism under defined conditions to study the relevance of proteins in a biological process.
NanoLC-MS plays an important role in both top-down and bottom-up proteomic analysis for protein identification. Depending on the sample complexity the set-up will comprise a single column or multiple columns for multidimensional separations.
LevelBasic
Proteomics aims to characterize the entire complement of proteins expressed in a cell, tissue, or organism under defined conditions to study the relevance of proteins in a biological process.
Samples are from biological origin, typically limited in amount and are very complex in composition. The sample complexity is the result of the large number of proteins present that ![]() differ in size, hydrophobicity, structure, and concentration.
differ in size, hydrophobicity, structure, and concentration.
In proteomics proteins are at some stage often converted to peptides by digestion with a specific enzyme such as ![]() trypsin:
trypsin:
- The digestion can be performed early in the sample preparation step without any protein pre fractionation. This analysis strategy is called bottom-up or shot-gun proteomics and yields very complex samples consisting of up to several thousand peptides.
- On the contrary, protein separation can be applied to some extend prior to the digestion step. This analysis strategy is called top-down proteomics.
Nano LC of peptides
Reversed phase ion-pairing nanoLC using packed or monolithic columns is well suited for the separation of peptides. Peptides are typically eluted with a binary solvent gradient consisting of water and acetonitrile and an ion-pairing agent. The gradient is normally started at 5% acetonitrile with a linear increase to 60% v/v. The ion-pairing agent masks the peptide charge and increases the hydrophobicity of the peptide yielding better retention on the reversed phase support.
Good results are obtained with tri-fluoroacetic acid and formic acid. Tri-fluoroacetic acid has a lower UV absorption at 214 nm and results in optimal chromatographic performance. Formic acid is preferred with ESI-MS detection as it often leads to better ionization efficiency.
The separation performance of a gradient LC system can be expressed by the ![]() peak capacity, i.e. the number of compounds that can be baseline separated in the gradient time.
peak capacity, i.e. the number of compounds that can be baseline separated in the gradient time.
The peak capacity can be increased by:
- Reduction of the stationary phase particle size
- Increasing the column length
- Extending the gradient time
- Optimizing the column temperature
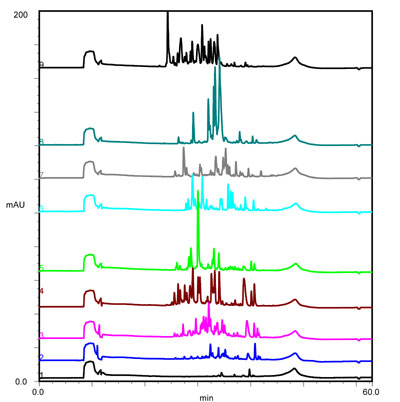
Separation of tryptic digest sample of bovine serum albumin at various gradient lengths. (Click for details)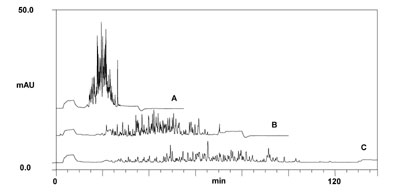
Peptide sequencing by nanoLC-MS/MS
The subject on peptide sequencing is also treated in the chapter on peptide fingerprinting. The workflow from sample to protein identification in a typical proteomic analysis consists of the following steps:
- Protein isolation and separation
- Protein digestion
- Peptide separation
- MS spectrum of peptide
- Fragmentation MS/MS spectrum of peptide
- Matching against database or manual interpretation
Proteins are isolated from the organism by biochemical fractionation. To reduce the complexity of the sample proteins are separated by (2D) gel electrophoresis or liquid chromatography. Despite recent progress in MS technology, e.g. gas phase fragmentation of proteins, MS on peptides is more easy and sensitive.
For that reason proteins are enzymatically converted to peptides. The most widely applied enzyme for this digestion step is trypsin which cleaves the protein at the carboxylic site of arginine and lysine. The resulting tryptic peptides that are protonated at the amino acid of the C terminus, are readily separated on reversed phase C18 columns and easily detected by MS detection.
The separation is preferably performed on a nanoLC column which is coupled to a tandem mass spectrometer. After solvent evaporation the (multiple) charged peptide is detected by MS yielding a peptide MS spectrum. In the last step of the analysis, selected peptides are isolated in the MS, fragmented by collision with an inert gas, and the MS/MS or fragmentation spectrum is ![]() recorded. The obtained MS/MS spectrum of the peptides giving information on the amino acid sequence of the protein, can be matched against databases to finalize identification. Reversed-phase nanoLC coupled to tandem mass spectrometry is a fast and sensitive method for peptide sequencing, which can be automated completely. This technique has matured into a standard tool for proteomics research.
recorded. The obtained MS/MS spectrum of the peptides giving information on the amino acid sequence of the protein, can be matched against databases to finalize identification. Reversed-phase nanoLC coupled to tandem mass spectrometry is a fast and sensitive method for peptide sequencing, which can be automated completely. This technique has matured into a standard tool for proteomics research.
Multidimensional nanoLC
A sample strategy with no or only limited protein pre-fractionation followed by enzymatic digestion renders very complex peptide mixtures. The number of peptides will, by far, exceed the peak capacity of a single reversed-phase nanoLC separation resulting in excessive co-elution. In particular low abundant peptides will not be detected and sequenced by the MS thereby potential important information can be missed. In addition to chromatographic optimization and using the selectivity of mass spectrometric detection (repeat the analysis with several non-overlapping restricted m/z ranges) a very powerful technique to improve the separation resolution is to use multidimensional chromatography.
In this technique the separation power is increased by adding a second separation dimension or selectivity to the nanoLC system. One of the most widely used multidimensional LC techniques for peptides is a 2D-LC method based on a strong cation exchange (SCX) and reversed phase column. At low pH (tryptic) peptides bear a positive charge assuring binding to the SCX column.
In the 2D-LC analysis
- A complex peptide mixture is first separated on an ion-exchange column.
- All the different peptide fractions eluting from the first dimension column are on-line injected and
- Separated on a second-dimension reversed-phase nanoLC column.
Several method parameters, such as column arrangement, column dimensions, salts for elution from the SCX column and the ion-pairing agent need to be considered in method ![]() development.
development.
A large variety of on-line and off-line 2D-LC techniques has been developed. While off-line techniques offer more method flexibility and the ability to re-analyse fractions from the first dimension separation, on-line technique are easy to set-up, provide full-automation, and are much faster. A commonly applied 2D-LC method is based on the set-up for sample pre-concentration. The set-up is extended with an SCX column which is eluted by performing a salt step gradient. The schematic is shown below.
Schematic 2D-LC salt steps (Click to open Power Point animation,Members only)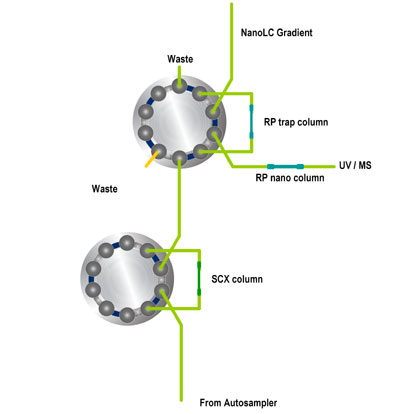 Steps in micro column switching:
Steps in micro column switching:
- Sample injection by autosampler onto SCX column
- Sample compounds that are not retained reach RP trap column
- Sample preconcentration takes place due to extremely high retention factors
- Valve switching to place RP trap column in series with RP nano column
- NanoLC gradient started to elute compounds from RP column
- Detection by UV and/or MS
- Valve switching to prepare for next injection (not shown)
- Injection of salt solution by autosampler to elute fraction of sample components (not shown)
An example of an on-line 2D-LC separation of a tryptic peptide sample is shown below.